Natural Products Research Summary
In the natural products part of the Clardy laboratory we are interested in finding new biologically active natural products, elucidating their modes of action, and understanding their biosynthesis. Following the hypothesis that greater genetic diversity of the producing organism leads to greater chemical diversity in the natural products produced, we explore extracts from a wide variety of organismal sources including endophytic fungi, insects, and higher plants. We also use the power of high-throughput screening at the ICCB to find natural products affecting a diverse array of interesting biological systems, and enjoy collaborations with many different biology labs in the Longwood Medical Area and beyond.
Following are some examples of natural products projects underway in the Clardy lab:
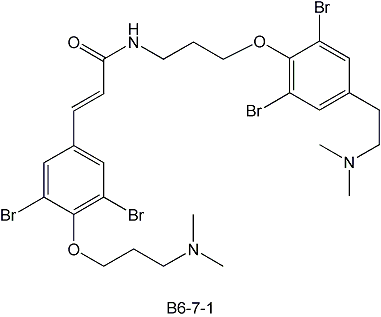
1. Identification of a PI3K/Akt/FOXO1a specific inhibitor from a marine sponge extract. In collaboration with Pam Silver?s lab at HMS, we identified the structure of the natural product B6-7-1, below, which inhibits PI3K/PTEN/Akt signaling. B6-7-1 was highly active in a cell-based, small molecule screen based on FOXO1a localization, and could prove to be helpful in the development of novel cancer therapies.
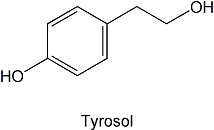
2. Isolation of a quorum-sensing molecule from Candida albicans. In collaboration with Gerald Fink's laboratory at the Whitehead Institute, our lab identified tyrosol as a quorum-sensing molecule in the yeast Candida albicans. Quorum sensing molecules are released by cells to monitor their population density and stimulate coordinated behavior when a threshold concentration of the molecule is reached. In the case of Candida albicans, the accumulation of tyrosol in the growth medium increases with increasing cell density. When added to dilute cultures of C. albicans, tyrosol reduces the lag phase of growth. Additionally, tyrosol stimulates germ-tube formation.
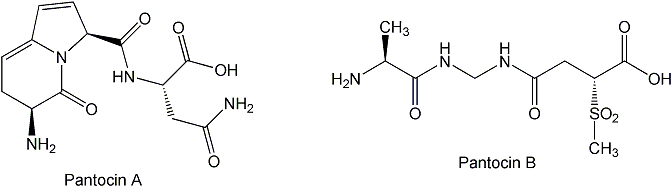
3. Isolation and biosynthesis of antibiotics from Pantoea agglomerans. In collaboration with Steve Beer's laboratory at Cornell University, we have been isolating antibiotics from the bacterium Pantoea agglomerans that are active against the plant pathogen Erwinia amylovora. P. agglomerans produces a diverse array of natural products and is a good model organism for studying biosynthetic diversity oriented synthesis. Using cosmid libraries of P. agglomerans DNA in E. coli, we have been able to isolate antibiotics from different strains of P. agglomerans and learn about their biosynthesis.
References
(1) Chen, H., Fujita, M., Feng, Q., Clardy, J., and Fink, G.R. "Tyrosol is a quorum-sensing molecule in Candida albicans" Proc. Natl. Acad. Sci. USA 2004, 101, 5048-5052.
(2) Kau, T.R., Schroeder, F., Wojciechowski, C.L., Ramaswamy, S., Zhao, J.J., Roberts, T.M., Clardy, J., Sellers, W.R., and Silver, P.A. "A chemical genetic screen for inhibitors of FOXO1a nuclear export" Clinical Cancer Research 2003, 9, 6138S-6139S.
(3) Jin, M., Liu, L., Wright, S.A.I., Beer, S.V., and Clardy, J. "Structural and functional analysis of pantocin A: An antibiotic from Pantoea agglomerans discovered by heterologous expression of cloned genes" Angew. Chem. Int. Edit. 2003, 42, 2898-2901.
(4) Jin, M., Wright, S.A., Beer, S.V., and Clardy, J. "The biosynthetic gene cluster of pantocin A provides insights into biosynthesis and a tool for screening" Angew. Chem. Int. Edit. 2003, 42, 2902-2905.